7 Groundwater
Purpose: derive metrics of groundwater influence for reporting unit locations
7.1 Data
Load spring prevalence rasters, lakes, basin and flowline
Code
# spring prevalence from Maxent: complete tiff and buffered/no lake tiff
spring_full <- rast("/Users/jeffbaldock/Library/CloudStorage/GoogleDrive-jbaldock@uwyo.edu/Shared drives/wyo-coop-baldock/UWyoming/Snake River Cutthroat/Analyses/Groundwater Seeps/SpringTIFFs/SpringPrev_SnakeGreys_BedSurf.tif")
# lakes in Upper Snake
lakes <- vect("/Users/jeffbaldock/Library/CloudStorage/GoogleDrive-jbaldock@uwyo.edu/Shared drives/wyo-coop-baldock/UWyoming/Snake River Cutthroat/Data/Spatial/Lakes/UpperSnake_Lakes.shp")
# basin without lakes
basin <- vect("/Users/jeffbaldock/Library/CloudStorage/GoogleDrive-jbaldock@uwyo.edu/Shared drives/wyo-coop-baldock/UWyoming/Snake River Cutthroat/Data/Spatial/Basin Delineation/BasinDelineation/MajorBasins_Watersheds.shp")
basin <- subset(basin, basin$site %in% c("UpperSnake", "Greys"))
basin_nolakes <- erase(basin, lakes)
# flowline
flowline <- vect("/Users/jeffbaldock/Library/CloudStorage/GoogleDrive-jbaldock@uwyo.edu/Shared drives/wyo-coop-baldock/UWyoming/Snake River Cutthroat/Analyses/Snake River GSI Quarto/Landscape Covariates/Watershed Delineation/SnakeGreys_flowline.shp")
flowbuff <- flowline %>% buffer(width = 100)Remove lakes and buffer to flowline
Write-out spring prevalence rasters
Code
writeRaster(spring_nolakes, "/Users/jeffbaldock/Library/CloudStorage/GoogleDrive-jbaldock@uwyo.edu/Shared drives/wyo-coop-baldock/UWyoming/Snake River Cutthroat/Analyses/Groundwater Seeps/SpringTIFFs/SpringPrev_SnakeGreys_BedSurf_nolakes.tif")
writeRaster(spring_nolakes_buff100, "/Users/jeffbaldock/Library/CloudStorage/GoogleDrive-jbaldock@uwyo.edu/Shared drives/wyo-coop-baldock/UWyoming/Snake River Cutthroat/Analyses/Groundwater Seeps/SpringTIFFs/SpringPrev_SnakeGreys_BedSurf_nolakes_flowbuff100.tif")Load spring prevalence rasters
Code
spring_nolakes <- rast("/Users/jeffbaldock/Library/CloudStorage/GoogleDrive-jbaldock@uwyo.edu/Shared drives/wyo-coop-baldock/UWyoming/Snake River Cutthroat/Analyses/Groundwater Seeps/SpringTIFFs/SpringPrev_SnakeGreys_BedSurf_nolakes.tif")
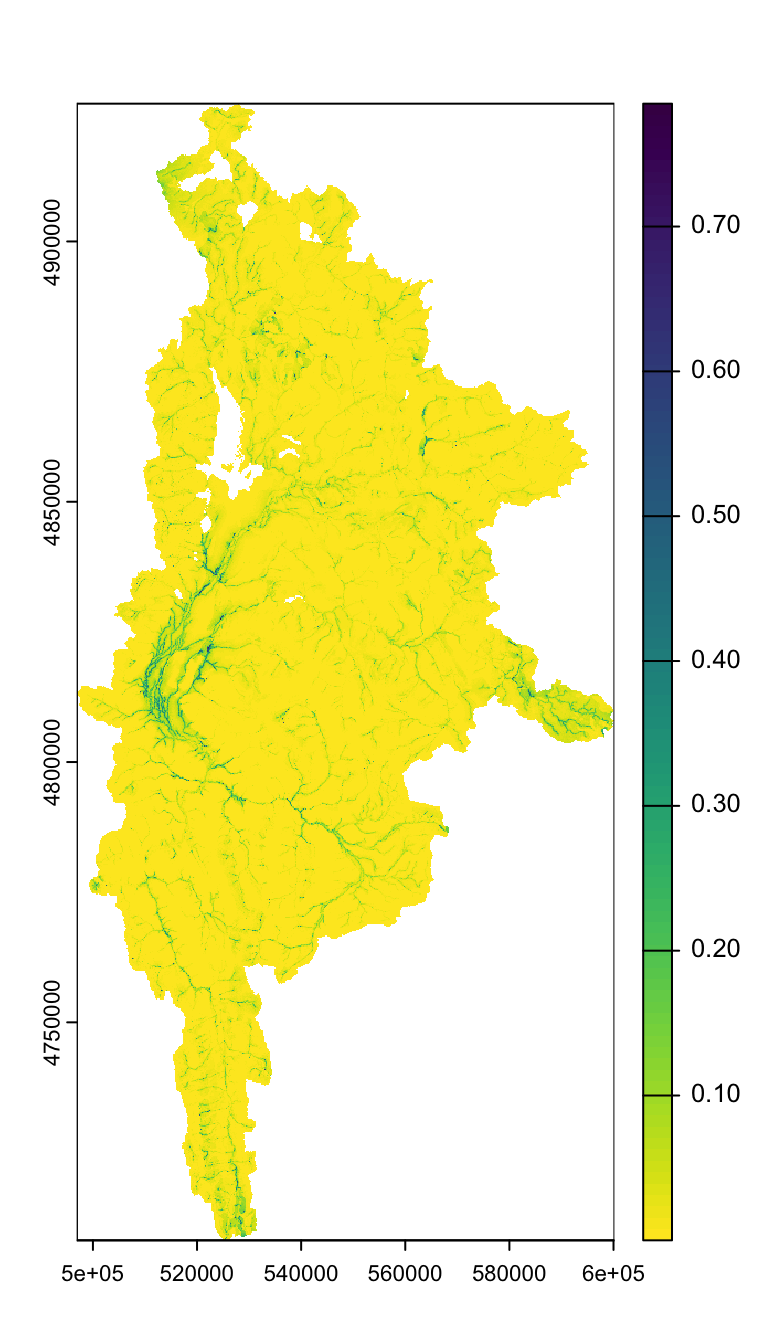
spring_nolakes_buff100 <- rast("/Users/jeffbaldock/Library/CloudStorage/GoogleDrive-jbaldock@uwyo.edu/Shared drives/wyo-coop-baldock/UWyoming/Snake River Cutthroat/Analyses/Groundwater Seeps/SpringTIFFs/SpringPrev_SnakeGreys_BedSurf_nolakes_flowbuff100.tif")View MaxEnt spring prevalnce raster
Buffered to flowline…
Code
plot(spring_nolakes_buff100)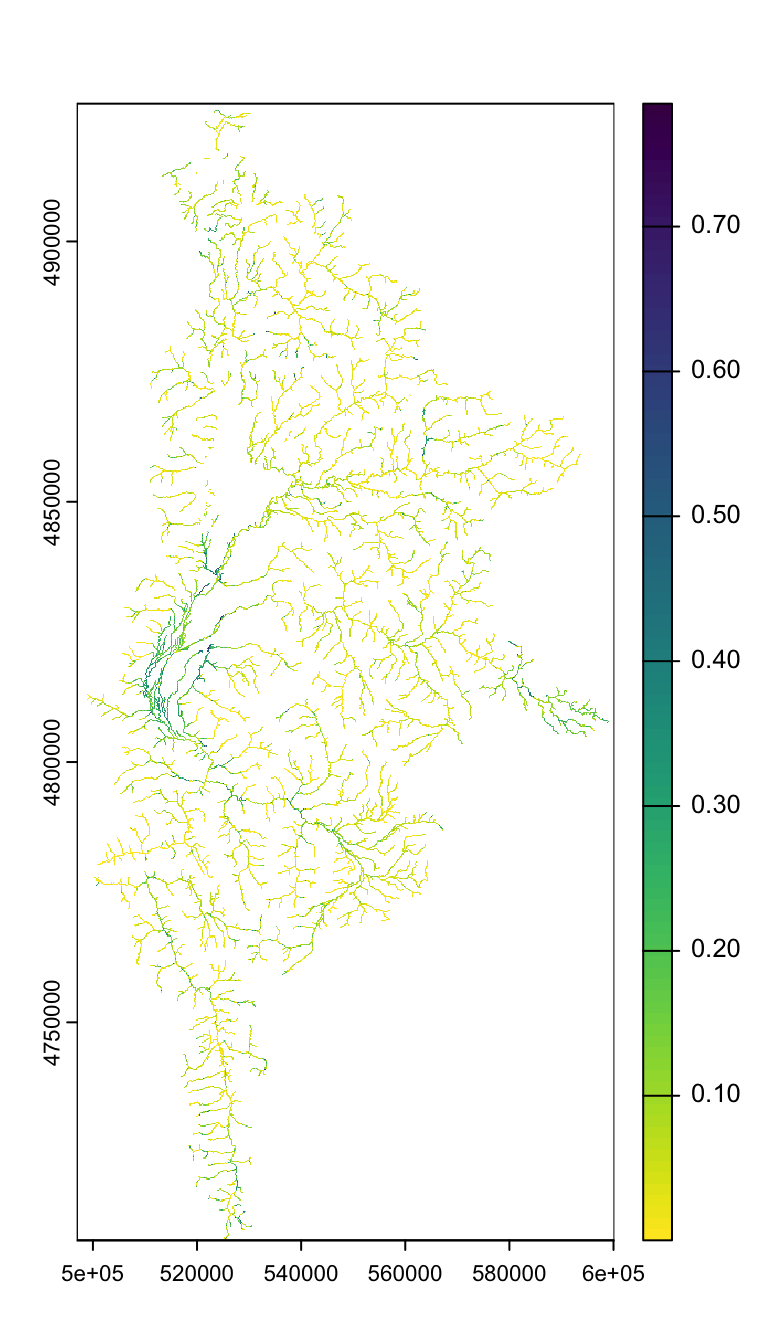
Reporting unit locations and watersheds
Code
# reporting unit locations
sites_repu <- vect("/Users/jeffbaldock/Library/CloudStorage/GoogleDrive-jbaldock@uwyo.edu/Shared drives/wyo-coop-baldock/UWyoming/Snake River Cutthroat/Analyses/Snake River GSI Quarto/Landscape Covariates/Location Data/RepUnit_SpatialLocations.shp")
# watershed shapefiles
sheds_repu <- read_sf(dsn = "/Users/jeffbaldock/Library/CloudStorage/GoogleDrive-jbaldock@uwyo.edu/Shared drives/wyo-coop-baldock/UWyoming/Snake River Cutthroat/Analyses/Snake River GSI Quarto/Landscape Covariates/Watershed Delineation", layer = "RepUnits_Watersheds")View reporting units and watersheds
7.2 Derive groundwater metrics
Calculate groundwater metrics for each basin. Extract average and weighted spring prevalence for each basin
Code
#
sites <- sheds_repu$site
gwlist <- list()
st <- Sys.time()
for (i in 1:length(sites)) {
spring_mask <- mask(crop(spring_nolakes_buff100, sheds_repu[sheds_repu$site == sites[i],]), sheds_repu[sheds_repu$site == sites[i],]) # crop and mask by basin
dist_rast <- distance(spring_mask, sites_repu[sites_repu$repunit == sites[i],]) %>% mask(spring_mask) # calculate distance between each raster cell and site location
gwlist[[i]] <- tibble(site = sites[i],
areasqkm = sheds_repu$aresqkm[i],
gwi_point = terra::extract(spring_nolakes_buff100, sites_repu[sites_repu$repunit == sites[i],], na.rm = TRUE)[,2],
gwi_iew05km = as.numeric(global(spring_mask * (1 / exp(dist_rast/5000)), "sum", na.rm = T) / global(1 / exp(dist_rast/5000), "sum", na.rm = T))
)
print(i)
}
Sys.time() - st
gwmetrics_repu <- do.call(rbind, gwlist) # bind as tibble
write_csv(gwmetrics_repu, "Landscape Covariates/Groundwater/GroundwaterMetrics_raw_RepUnits.csv")Code
| site | areasqkm | gwi_point | gwi_iew05km |
|---|---|---|---|
| bailey_NA | 41.5714362 | 0.0095202 | 0.0281587 |
| blackrock_lower | 124.9215920 | 0.1393530 | 0.0438536 |
| blackrock_upper | 80.6862046 | 0.1924090 | 0.0685266 |
| blacktail_NA | 61.1445506 | 0.1783010 | 0.2664975 |
| blindbull_NA | 36.3517449 | 0.0473298 | 0.0702745 |
| boulder_NA | 53.6999801 | 0.1402050 | 0.0497372 |
| box_NA | 29.1414701 | 0.0594441 | 0.0251458 |
| cabin_NA | 23.5100427 | 0.0558411 | 0.0497856 |
| clear_NA | 16.6041016 | 0.0753176 | 0.0287769 |
| cliff_NA | 158.3813174 | 0.2303060 | 0.0598667 |
| cody_bluecrane | 18.5117071 | 0.5193600 | 0.2969744 |
| cottonwood_grosventre | 89.1780292 | 0.1426020 | 0.0742731 |
| cottonwood_nps | 187.1527021 | 0.4438450 | 0.1516091 |
| cowboycabin_NA | 6.6392953 | NA | 0.1510349 |
| crystal_lower | 185.1148874 | 0.0840567 | 0.0534595 |
| crystal_upper | 155.6115004 | 0.0156552 | 0.0350789 |
| deadman_greys | 42.6069634 | 0.0568219 | 0.0415624 |
| deadmansbar_NA | 11.7612083 | 0.4607530 | 0.1228488 |
| dell_NA | 118.1568591 | 0.2145850 | 0.0506646 |
| ditch_NA | 67.6914863 | 0.0331338 | 0.0379450 |
| dog_NA | 31.0904837 | 0.0636842 | 0.0618599 |
| fall_coburn | 116.5315084 | 0.2628070 | 0.1059285 |
| fish_NA | 233.3741441 | 0.1869660 | 0.1932733 |
| fish_grosventre | 587.8758585 | 0.0790754 | 0.0570004 |
| flat_NA | 173.3686411 | 0.4952880 | 0.2374473 |
| fordspring_NA | 0.3712985 | 0.3635020 | 0.3083826 |
| goosewing_NA | 40.3960756 | 0.1272550 | 0.0449844 |
| granite_lower | 220.3826608 | 0.4365610 | 0.0792698 |
| granite_upper | 131.1747886 | 0.0685593 | 0.0512103 |
| grosventre_lower | 1589.6465468 | 0.1128500 | 0.1277220 |
| hoback_upper | 114.0778330 | 0.1322940 | 0.0672652 |
| horse_NA | 63.2035547 | 0.3199460 | 0.0541071 |
| lava_NA | 65.3337290 | 0.1542410 | 0.0371282 |
| leidy_NA | 10.7360331 | 0.0376180 | 0.0454852 |
| littlegreys_steer | 179.3457294 | 0.1819260 | 0.0565315 |
| lowerbarbc_NA | 6.3968311 | 0.3782680 | 0.2384035 |
| mosquito_NA | 61.6412867 | 0.5507170 | 0.0881280 |
| northbuffalofork_NA | 211.7277107 | 0.4812010 | 0.1090835 |
| pacific_NA | 415.0574929 | 0.1369520 | 0.0415821 |
| rock_NA | 11.9857990 | 0.0256445 | 0.0192197 |
| shoal_NA | 82.6496949 | 0.1540520 | 0.0655054 |
| slate_NA | 97.2568238 | 0.0359981 | 0.0333669 |
| snakeriversidechannel_NA | 18.8103796 | 0.5644690 | 0.3722617 |
| spread_southfork | 98.9032021 | 0.0189719 | 0.0327670 |
| spread_uppermainstem | 202.6088599 | 0.0736471 | 0.0452715 |
| spreadnf_flagstaff | 62.1057535 | 0.0557931 | 0.0769342 |
| spring_nps | 4.0130319 | 0.2528840 | 0.0921442 |
| spring_tss | 24.8578294 | 0.3593580 | 0.2004281 |
| threechannel_NA | 32.3647548 | 0.2897590 | 0.1178438 |
| upperbarbc_NA | 5.7370505 | 0.4632450 | 0.3517652 |
| white_NA | 32.6721618 | 0.1206510 | 0.0640540 |
| willow_NA | 185.7033361 | 0.3712950 | 0.0722065 |